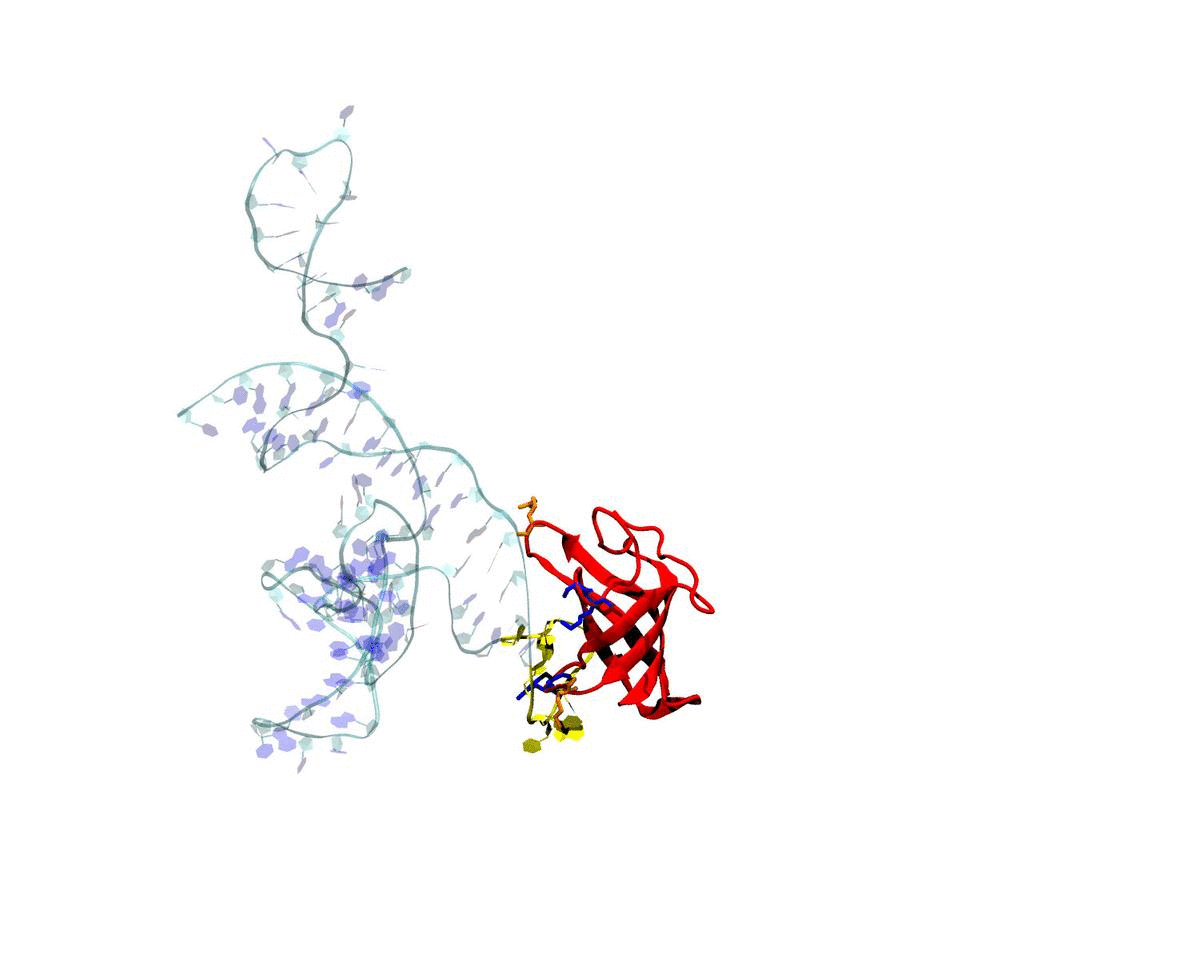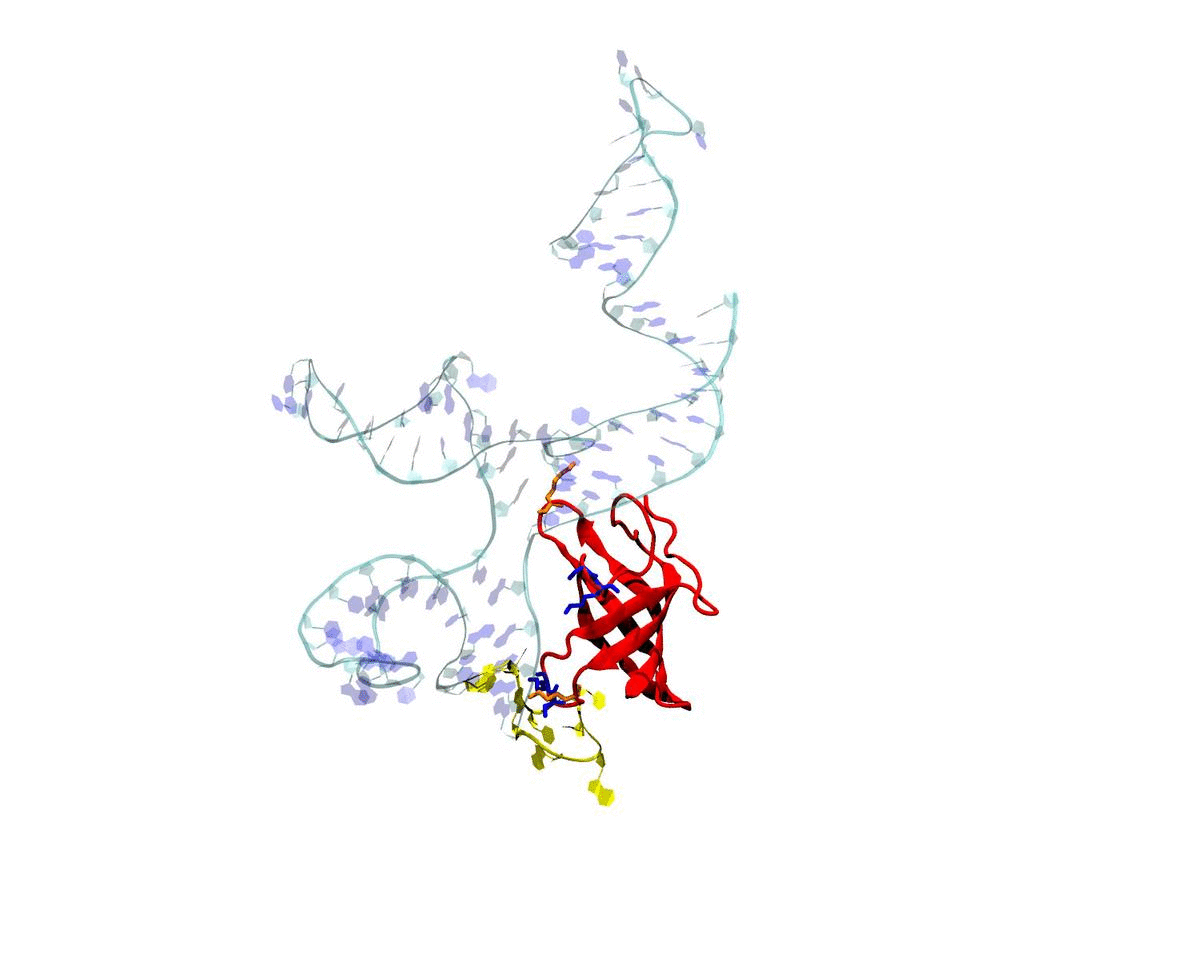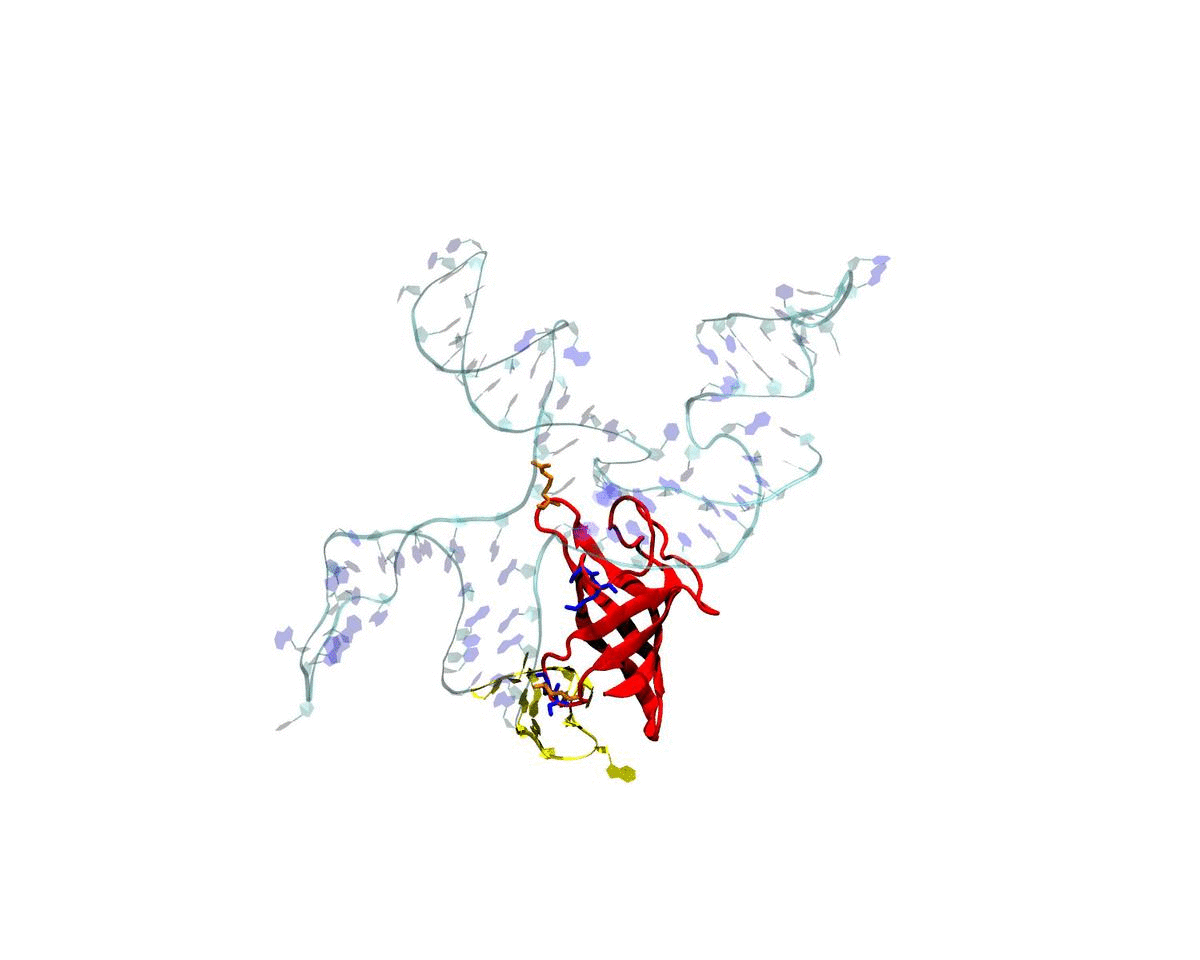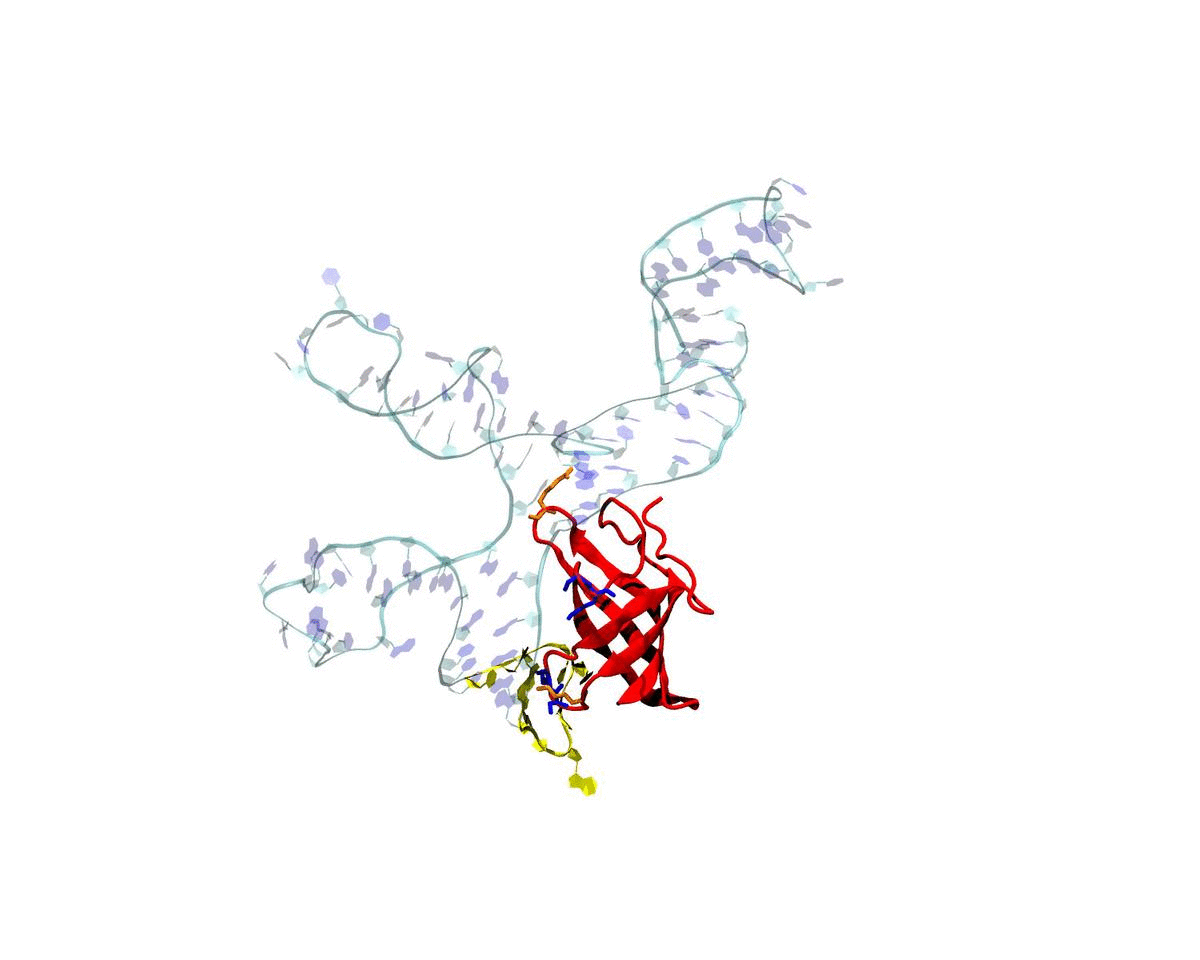
Introduction
RNA molecules can adopt stable secondary and tertiary structures, which are essential in mediating physical interactions with other partners such as RNA binding proteins(RBPs) and in carrying out their cellular functions. In vivo and in vitro experiments such as RNAcompete and eCLIP have revealed in vitro binding preferences of RBPs to RNA oligomers and in vivo binding sites in cells. Analysis of these binding data showed that the structure properties of the RNAs in these binding sites are important determinants of the binding events; however, it has been a challenge to incorporate the structure information into an interpretable model. Here we describe a new approach, RNANetMotif, which takes predicted secondary structure of thousands of RNA sequences bound by an RBP as input and uses a graph theory approach to recognize enriched subgraphs. These enriched subgraphs are in essence shared sequencestructure elements that are important in RBP-RNA binding. To validate our approach, we performed RNA structure modeling via coarse-grained molecular dynamics folding simulations for selected 4 RBPs, and RNA-protein docking for LIN28. The simulation results, e.g., solvent accessibility and energetics, further support the biological relevance of the discovered network subgraphs.